Feb. 10, 2021
 LA JOLLA, CA – The Center for Viral Systems Biology (CViSB) at Scripps Research is tracking the prevalence of new strains of SARS-CoV-2, the virus behind the COVID-19 pandemic. The team behind Outbreak.info has created a dashboard for viewing daily reports that describe the current situation, focusing on the United States, and has made this publicly available at https://outbreak.info/situation-reports.
LA JOLLA, CA – The Center for Viral Systems Biology (CViSB) at Scripps Research is tracking the prevalence of new strains of SARS-CoV-2, the virus behind the COVID-19 pandemic. The team behind Outbreak.info has created a dashboard for viewing daily reports that describe the current situation, focusing on the United States, and has made this publicly available at https://outbreak.info/situation-reports.
As concerns surrounding several COVID-19 variants have been developing, researchers around the world have shifted focus in an all-hands-on-deck, round-the-clock effort to uncover more about the mutations. Concerns are growing because these mutations have shown to be significantly more transmissible than other variants. This includes the B.1.1.7 lineage, also known as Variant of Concern 202012/01 (VOC-202012/01) or 20B/501Y.V1. This mutation was first identified in the UK in early December 2020 and has since been detected in the US and other counties.
In investigating emerging strains of SARS-CoV-2, one tool has been missing from the toolkit – a standardized hub of information about these mutations. To stay updated on the information needed to fight the pandemic, it is imperative that researchers have access to open source tools. It is vital to an informed citizenry that the public have access to open data and resources, as well. That is why the team decided to build a dashboard scientists and other individuals can use to find an easily accessible, reliable source of aggregated information that is updated daily.
Alaa Abdel Latif, Research Programmer on the CViSB team, says the tool was initially built to analyze the mutations found in their lab’s sequencing samples. “But given the recent concerns regarding emerging SARS-CoV-2 variants,” Latif shared, “we started applying it to analyze mutations found across the United States.”
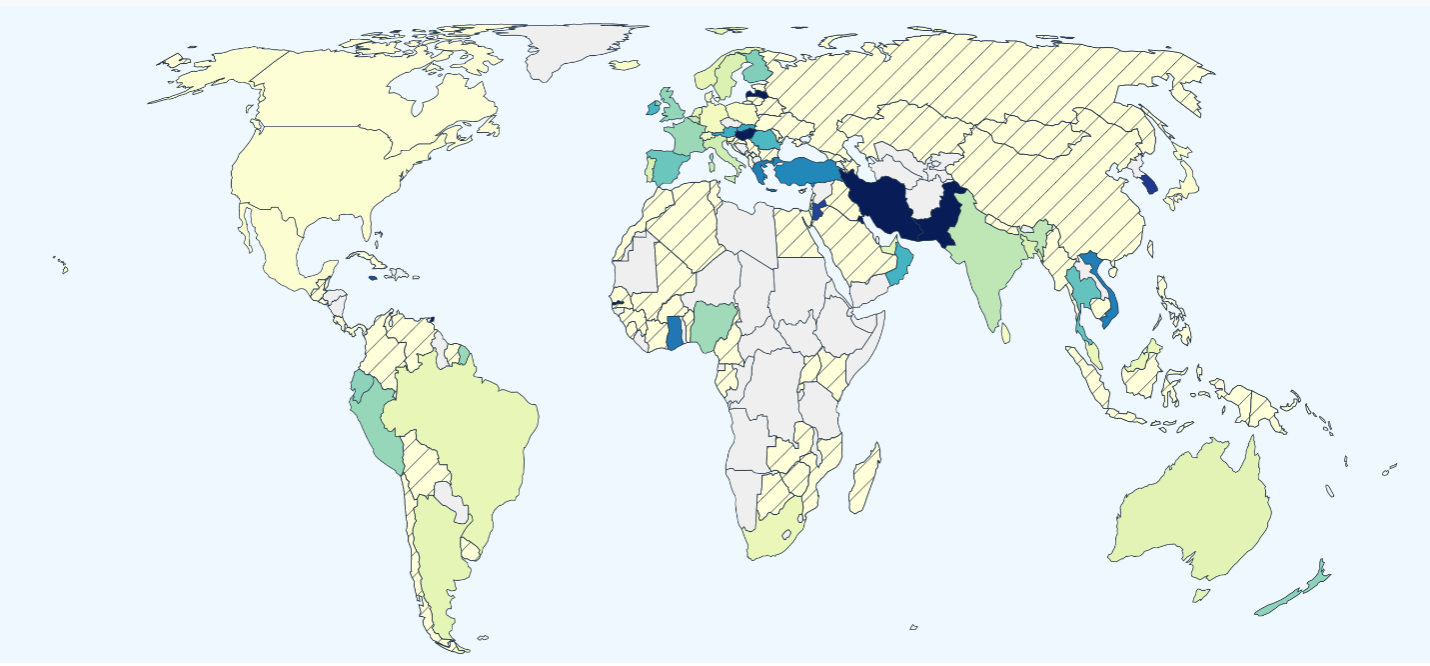
The dashboard includes daily situation reports for mutations and lineages (series of mutations). Using data generated from the GISAID Initiative, these reports track the number of sequences detected, locations where sequences have been detected, and prevalence over time at global, national, and state levels. The dashboard also includes mutation maps for easy comparison between lineages.
“Thanks to global scientific efforts, there are an unprecedented number of viral genomes of SARS-CoV-2 being produced and deposited to the GISAID database every day,” said Karthik Gangavarapu, doctoral student and computational scientist with CViSB. Gangavarapu says this was a natural extension of Outbreak.info, as it is important that the tool is scalable, updated daily, and allows analysis of these mutations over time.
The SARS-CoV-2 Mutation Situation Reports interface gives the world a way to track strains of the virus and stay updated on COVID-19 changes as they occur. Moreover, it gives researchers an advantage in breaking ground in the COVID-19 pandemic.
About the Center for Viral Systems Biology
The Center for Viral Systems Biology uses genomics, bioinformatics, computational biology, and other areas of research to investigate attributes of pathogenic viruses that determine the outcomes of human disease. The research center is located in Scripps Research in La Jolla, California and is supported by the National Institute of Allergy and Infectious Diseases, part of the National Institutes of Health (5U19 AI135995-03). For more information: https://cvisb.org/
About Outbreak.info
Outbreak.info was developed by the Andersen, Su, and Wu labs at Scripps Research in La Jolla, California. It was created to aggregate and standardize COVID-19 and SARS-CoV-2 data in a single tool that allows researchers to visualize and download epidemiology data or search for publications, datasets, clinical trials, and more. This is supported with funding from the National Institutes for Health (3U19AI135995-03S2). For more information: https://outbreak.info/
Contact: Emily Haag, 562-480-2021, [email protected]