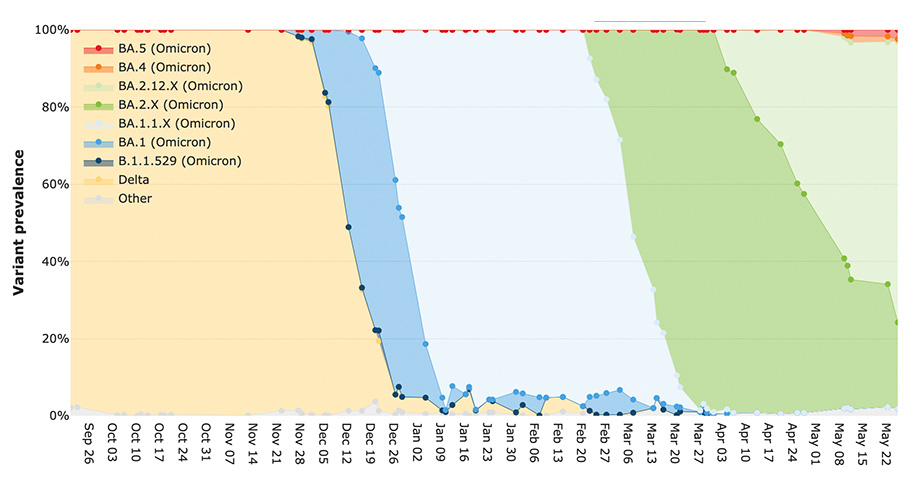
The CViSB developed tool, Freyja, is helping scientists detect SARS-COV-2 variants circulating in a community before they appear in clinical tests. As described in the recent Nature publication describing the study, wastewater sampling is a cheaper, faster, and more accurate way for public health officials and researchers to detect rising cases, and now, it can accurately determine the genetic mixture of SARS-CoV-2 variants present within a population and identify new variants of concern up to 14 days before traditional clinical testing.
“In a lot of places, standard clinical surveillance for new variants of concern is not only slow but extremely cost-prohibitive,” says Kristian Andersen, PhD, professor of Immunology and Microbiology at Scripps Research and a senior author of the new work. “But with this new tool, you can take one wastewater sample and basically profile the whole city.”
CViSB postdoc Josh Levy developed a library of “barcodes” that identify SARS-CoV-2 variants based on short snippets of their RNA that are unique to each variant. Then, he coded a new computational tool that sifts through the mass of genetic information in wastewater to find these barcodes. He made the new Freyja program easy to use and free.
“If you’re in a lab that can already sequence a wastewater sample, you’re good to go— you just run this code and in another 20 seconds you’re done,” he says.
When the researchers applied Freyja to their wastewater samples and compared the results to clinical data collected from around San Diego, they discovered that the tool detected variants of concern, including Alpha, Delta, and Omicron, in wastewater up to 14 days before it was reported clinically.
The researchers say they are continuing to improve upon the set of tools they use to analyze viruses in wastewater, but that the current suite of methods is already a leap forward from previous approaches. The same strategies could be used to not only track variants of SARS-CoV-2 but other human pathogens. Read more here.