Over the course of the COVID-19 pandemic, the research community has responded by sharing an unprecedented amount of data publicly, including millions of SARS-CoV-2 sequences, protein crystal structures, and in vitro assays which have allowed us to rapidly develop new diagnostics, treatments, and vaccines. Despite the widespread enthusiasm of the research community to share data publicly, however, disseminating and finding data efficiently across disparate networks has been a formidable task. In an effort to push through the barriers to effective data dissemination, a consortium focused on infectious disease data presented solutions in two recently published articles in Scientific Data, Developing a standardized but extendable framework to increase the findability of infectious disease datasets and Addressing barriers in FAIR data practices for biomedical data.
The NIAID/DMID Systems Biology Consortium for Infectious Diseases Data Dissemination Working Group (DDWG) is a group of bench scientists, bioinformaticians, computational biologists, and clinicians united by their goal to develop practical solutions to make their data more findable, accessible, interoperable, and reusable (FAIR). The DDWG draws from current and previous members of the Systems Biology Consortium for Infectious Diseases Centers, a collaborative network established by the National Institute of Allergy and Infectious Diseases (NIAID) which includes the Center for Viral Systems Biology (CViSB). These groups collectively are providing insights into the relationships between humans and pathogens using a Systems Biology approach that combines several interdisciplinary techniques together.
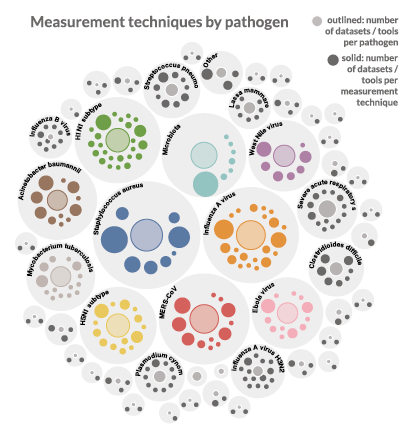
The Centers within the SysBio groups generate volumes of data and software that are shared publicly, but finding these resources deposited in different repositories can be challenging. To tackle these problems, the DDWG analyzed current standards across data repositories and identified that there was a wide variation in how repositories categorized and provided access to datasets. For data to be easily findable, data needs to be described with comprehensive descriptions, called metadata. Metadata is standardized when mapped to a common schema, which defines the different elements of data and the rules for using descriptions. By creating a single set of standards for structuring metadata, search engines can find data.
The DDWG developed the NIAID SysBio schemas, reusable metadata schemas based on the widespread Schema.org standard, and registered datasets and computational tools created by the centers. These schemas link datasets to their funding information, experiment type, pathogen, host, and other pieces of metadata – increasing the findability of nearly 400 datasets and computational tools across the 15 research centers, regardless of which repository they are stored in or the type of data they contain. The SysBio schemas establish a standardized approach designed to be interoperable with community standards and customized for biomedical application. The developed schemas are shared openly to promote widespread use beyond the NIAID data ecosystem in hopes they can be used to mitigate FAIR data problems, support data discovery, and accelerate research within the broader scientific community.
Already, the NIAID SysBio schemas have been used in a variety of other infectious disease data sharing projects, including the Center for Viral Systems Biology Data Portal, the outbreak.info Research Library, and an immune-mediated and infectious disease Discovery Portal (NIAID Data Ecosystem).
As a result of ongoing collaborative effort across the Systems Biology Centers, barriers that have stood in the way of data reusability and scientific progress are being dismantled, but these are just the first steps. Based on their experiences increasing the findability of their own data, the DDWG also identified three ongoing challenges with sharing data and offered potential solutions to these barriers in an associated commentary. These solutions include developing long-term incentives for researchers to share data well, standardizing the creation of and sharing of metadata, and coordinating international data sharing efforts alongside sustained support of data tools and platforms. These recommendations come at a critical juncture as the scientific community tries to remodel its data sharing practices. At the beginning of the year, the NIH updated their data sharing policy to require scientists to share data as a condition of their research. This work helps researchers navigate the complicated logistics of data sharing with a practical guide to make the process as easy and effective as possible.
Making data easier to find increases the likelihood that diverse datasets will be integrated in novel analyses that shed new light on the mechanisms of infectious diseases. It also means that investigators who did not generate the data will be more likely to find it and perform secondary analyses or meta-analyses. With the standardized, maintained, and reusable model implemented by the DDWG, scientists can overcome data sharing and data finding obstacles, increasing the availability of their research for use by the wider scientific community.
This work was supported in part by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) grants U01 AI124290 (Baylor), U01 AI124302 (Boston College), U19 AI135995 (Scripps Research), U19 AI135964 (Northwestern), U19 AI135972 (Sanford Burnham Prebys); U01 AI124319 (UCLA), 75N91019D00024 (Scripps Research); National Center for Advancing Translational Sciences NIH grant U24 TR002306 (Scripps Research); and National Institute of General Medical Sciences grant R01 GM083924 (Scripps Research).