CViSB is 5 years old as of this February! Looking back, we have made significant strides toward our goal of identifying the human and viral factors that determine the outcome of infection with Lassa, Ebola, and SARS-CoV-2 viruses.
CViSB’s multi-disciplinary team has collected clinical, immunological, and genomic data, combined multi-omics technologies and applied high-throughput (HTP) experiments to our unique patient and survivor cohorts of Lassa fever in Sierra Leone, Ebola virus disease in Sierra Leone and the Democratic Republic of Congo (DRC), and COVID-19 in Sierra Leone and the United States.
Through the efforts of CViSB’s Technology, Data Management, and Modeling cores, data integration pipelines were developed along with novel analytical tools and techniques that allowed us to apply a Systems Biology approach to the analysis and interpretation of disparate datasets.
As a result, our team of researchers at Scripps Research, Tulane University, UCLA, MIT, Massachusetts General Hospital, and Kenema Government Hospital achieved a remarkable number of accomplishments and milestones. These include the publication of over 125 research articles describing the findings from data housed in the CViSB data portal. The data portal contains data from over 62,000 patients and 60,000 experiments. Our open-source database of SARS-CoV-2 variant data and COVID-19 epidemiological data, Outbreak.info, has driven research efforts by making vital information publicly available, including 13.5+ million SARS-CoV-2 sequences.
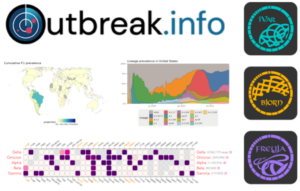 Additionally, CViSB researchers have developed 14 widely-used software packages and tools, which have been downloaded more than 3 million times. These include tools utilized by the FDA and CDC, such as Frejya, which estimates the relative abundance of SARS-CoV-2 lineages in wastewater samples. It also includes software packages like iVAR, used to assemble virus genomes, and Bjorn, which generates information for large-scale genomic surveillance of SARS-CoV-2 sequences.
Additionally, CViSB researchers have developed 14 widely-used software packages and tools, which have been downloaded more than 3 million times. These include tools utilized by the FDA and CDC, such as Frejya, which estimates the relative abundance of SARS-CoV-2 lineages in wastewater samples. It also includes software packages like iVAR, used to assemble virus genomes, and Bjorn, which generates information for large-scale genomic surveillance of SARS-CoV-2 sequences.
CViSB supported numerous high school, undergraduate and graduate students through matriculation and graduation, and more than 15 postdocs in their early careers as scientists! Our PIs strongly believed that participation in the CViSB Center gave these trainees valuable insight into how they can fit their experimental or computational expertise into cross-disciplinary approaches for infectious disease research.
Lastly, CViSB contributed to scientific research capacity building, particularly in West Africa, through bioinformatics training in the Democratic Republic of Congo, advanced technology and protocol training in Sierra Leone and hosting Sierra Leonean students at both Tulane University and Scripps Research. We also hosted two onsite systems biology and bioinformatics workshops (and one joint virtual research symposium) that were so well received that similar CViSB workshops will be offered again in the upcoming years.
Given these many accomplishments, we are thrilled that our forward momentum will be sustained with the renewal of CViSB’s funding support from the National Institutes of Health (NIH) for an additional 5 years. The next phase of CViSB will build on the well-laid foundation established by CViSB’s stellar team. We will expand CViSB’s systems biology analyses of host, virus, and environmental factors that contribute to the severity and spread of human diseases. We will accomplish this through the generation of new Lassa, Ebola and SARS-CoV-2 datasets, open-source tools and resources, as well as translational approaches.
Many thanks to the wonderful team of scientists past and present who have contributed to CViSB’s success. We look forward to the new research, resource development, and opportunities the next phase of CViSB will bring.
